To determine the pattern of inheritance for a particular mutation, click on the Full Pedigree tab at the top of the screen. The pedigree that is displayed on this page if one of 100 different sample pedigrees for ALS available in PedigreeLab. To choose a different pedigree, click on one of up or down arrows in the upper right corner of the screen. The purpose of this Full Pedigree function is to provide you will a large number of different pedigrees that you can evaluate to develop a hypothesis for whether a particular mutation is inherited by autosomal recessive, autosomal dominant, or sex-linked inheritance.
- When studying these pedigrees, remember that circles represent females and squares represent males. Parents are connected by a red horizontal line. Black vertical lines from the parents indicate the offspring produced by these parents. Sibs are connected to each other by a horizontal black-lined bracket shown above the symbols. The symbol for an individual that expresses the mutant phenotypes is shaded blue. First generation parents are shown at the top of the pedigree followed by the second generation, third generation etc.
- You may already know that ALS is inherited as an autosomal dominant condition. Heterozygotes develop the disease while homozygous recessive individuals are phenotypically normal. Homozygous dominant individuals rarely live to adulthood because of the lethality of this mutation. Study several of the full pedigrees until you are comfortable with the inheritance of ALS as an autosomal dominant trait.
- Assigning Genotypes to the Individuals in a Pedigree
Select a simple pedigree with a first generation parent that has ALS and a first generation parent who is phenotypically normal. You can assign genotypes to each of the members of a pedigree as follows:- To assign a genotype to each parent, double click on the circle for the first generation mother. A new box should open prompting you to type in the mother's genotype. Type in "Aa" or "aa" depending on whether the mom is a heterozygote with ALS or a phenotypically normal homozygote, then click OK to close the genotype box. Double click on the square for the first generation father and type in the appropriate genotype for the father, then click OK to close the genotype box. The genotype for each parent should appear below each symbol.
- Repeat this process to assign genotypes for the second and third generation individuals in the pedigree. Once you have filled in the genotypes for a pedigree. The labeled pedigree can be exported as a GIF file by clicking on the Export Pedigree button at the lower right corner of the screen. A new browser window with your labeled pedigree will appear. You can now print your pedigree from this window. Analyze at least two other pedigrees to confirm your genotype assignments.
- To assign a genotype to each parent, double click on the circle for the first generation mother. A new box should open prompting you to type in the mother's genotype. Type in "Aa" or "aa" depending on whether the mom is a heterozygote with ALS or a phenotypically normal homozygote, then click OK to close the genotype box. Double click on the square for the first generation father and type in the appropriate genotype for the father, then click OK to close the genotype box. The genotype for each parent should appear below each symbol.
- Mapping the Locus for a Mutant Gene by Studying Large Family Pedigrees, RFLP Analysis, and Recombination
Identifying the chromosome that contains the mutation that you are studying and mapping the locus for this mutation using PedigreeLab involves your ability to apply what you learned about the mode of inheritance for the mutation, and your ability interpret the results of RFLP and recombination data. This process is part of the challenge and fun of PedigreeLab.
Using the Large Family function is the key to the gene mapping functions of PedigreeLab. This function simulates databases that present family pedigrees for individuals with different genetic disorders that have been subjected to RFLP analysis using one of 17 different probes for markers on human chromosomes. The pedigrees are arranged according to genotypes for the RFLPs; phenotypes are also shown with these pedigrees. The probes in PedigreeLab do not correspond to actual marker loci in human chromosomes.
You will then pick an appropriate pedigree that will allow you to look for recombination between the marker sequence of DNA recognized by the probe and the mutation. Based on recombination frequencies that you generate, and LOD scores, you will then decide whether the probe and mutation are linked to the same chromosome. If they are linked, you will then use the Chromosome View function to pinpoint the locus for your trait based on recombination frequencies. All of the traits presented in PedigreeLab will map to their actual chromosomes as found in real life. No shortcuts or hints are given to determine linkage groups, hence you must be a gene hunter using trial and error and analysis of your data to eliminate loci for unlinked probes and find linked probes that will help you pinpoint the locus for the mutant gene!
- Begin by clicking on the Large Family tab at the top of the screen. Before you view the family pedigrees, you are asked to answer three questions on the expression of the mutant phenotype in a pedigree. You have the option of selected whether the mutant trait is expressed in grandparents, parents, and offspring. The purpose of these three questions is to better define and narrow your search of the Large Family databases to select for those pedigrees that will be most helpful to study recombination.
- Remember that for most traits you will want to define the search to look for pedigrees that have one parent who is a double heterozygote for the trait and probe (marker) and one parent who is a double homozygote for the trait and probe.
- For this ALS assignment, answer "yes" to each question. For the other assignments that you will be working on, think carefully about the mode of inheritance of the trait that you are working on before answering yes or no to each of the three questions. Before searching the database. Note that the default probe selected should be the "asr" probe. Click on the Search Database button. Your search of the database will reveal different family pedigrees that are assigned genotypes resulting from RFLP analysis with the asr probe.
- Remember that for most traits you will want to define the search to look for pedigrees that have one parent who is a double heterozygote for the trait and probe (marker) and one parent who is a double homozygote for the trait and probe.
- RFLP Analysis
The first screen that appears after searching the Large Family database displays a pedigree in which individuals are labeled according to their phenotype expressed and assigned a genotype according to the results of RFLP analysis with the asr probe.- To interpret each phenotype, note that the symbols for individuals expressing the mutant phenotype are shaded in blue. You can assign a genotype for the trait to each individual in the pedigree by double clicking on each symbol as you did with the Full Pedigrees and entering the genotypes for each individual in the pedigree.
- To interpret the results of the RFLP analysis, genotypes are labeled in each symbol of the pedigree according to the presence (+) or absence (-) of a restriction site in the fragments of DNA detected by the probe. Individuals who are homozygous for the presence of a restriction site are labeled as "+/+." Heterozygous individuals are labeled as "+/-", indicated the absence of a restriction site on one homologue and the presence of a restriction site on the other homologue. Homozygous individuals lacking a restriction site on both homologues of a pair are labeled as "-/-."
- Notice that an autoradiogram showing the RFLP pattern that would appear for different individuals depending on their RFLP genotype is shown on the right side of the screen. Reading the autoradiogram from left to right, the first lane, labeled "Ref," consists of a series of DNA fragments of known size in kilobases (kb) that are used as size standards or references for determining the size (in base pairs) of the RFLP fragments. Clicking on the symbol for any individual in the pedigree will show their RFLP pattern in the "selection" lane of the autoradiogram. Compare the size of each DNA fragment to the size standards shown in the far left lane; note that large DNA fragments are located at the top of the autoradiogram while smaller fragments migrate relatively faster towards the bottom of the autoradiogram. You may want to refer back to figure 1 in the background section of this lab to help you interpret the RFLP autoradiograms.
- To interpret each phenotype, note that the symbols for individuals expressing the mutant phenotype are shaded in blue. You can assign a genotype for the trait to each individual in the pedigree by double clicking on each symbol as you did with the Full Pedigrees and entering the genotypes for each individual in the pedigree.
- Choosing a Large Family Pedigree to Study Recombination Between the Trait and Marker
Once you are comfortable with how a Full Pedigree is shown, use the arrows in the upper right corner of the screen until you see a pedigree that is appropriate for studying recombination. You should begin by looking for a pedigree with one parent who is a double heterozygote (A/a, +/-) for the trait and probe, and a parent who is a double homozygote for the trait and probe (a/a, +/+). Look carefully at the phenotype and RFLP genotype of the grandparents before assigning genotypes to the parents. Once you have found an appropriate pedigree, you may find it helpful to label all of the individuals in the pedigree before attempting to count recombinant and nonrecombinant offspring. Hint: If you are having difficulty finding the correct pedigree, use the Genetic Calculator function described below.- The Genetic Calculator function can be very useful for helping you determine which pedigrees will be best suited for studying recombination between the trait and marker. This function allows you to specify genotypes in the parents of a full pedigree that show the trait and marker linked to a pair of homologous chromosomes or unlinked on different chromosomes.
To use this function, click on the Genetic Calculator tab at the top of the screen, then click on the popup arrow next to the box labeled "Trait & Probe Autosomal, Linked" at the top of this window. This popup menu allows you to design genotypes for different individuals depending on whether you think that the trait you are trying to map is an autosomal or sex-linked trait, and whether or not this trait is linked or unlinked to the probe for a given marker. Because you know from studying the full pedigrees that ALS is inherited as an autosomal dominant condition, select the "Trait & Probe Autosomal, Linked" to design genotypes that would show the ALS trait and asr marker as linked on an autosome.
Notice that on the far left of this box you have four popup menu boxes that you can use to select the trait and marker combination for each homologue of a pair in a female parent. Click on one of the upper two boxes, these allow you to choose the genotype for the trait. Note that the dominant allele for ALS is shown in capital letters and the recessive allele for ALS is shown in lower case letters. Click on one of the lower two boxes, these allow you to choose the genotype for the probe (marker). Note that the genotype for a homologue with a restriction site is shown as "asr+", and the genotype for a homologue lacking a restriction site is shown as "asr-."
It is important that you determine the correct linkage combination for the parent who is a double heterozygote (A/a, +/-) for the trait and restriction site. Do this for the female parent. For the first homologue select a linkage combination with "ALS" and "asr+" on the first homologue. Select the opposite homologue with the linkage configuration of "als" and "asr-."
Repeat this process for a male parent who is a double homozygote for the trait and restriction site (a/a, +/+). For this parent, the linkage combination is the same on both homologues.
Once you have selected the genotypes for each parent, click on the Calculate Punnett Squares button. You should now see Punnett squares that show nonrecombinant and recombinant genotypes that would result from a cross between the parents with the trait and restriction site arrangements that you just set up. You are now ready to return to the Large Family pedigree function to look for a pedigree with parents that show the genotypes for the trait and marker combination that you set up with the Genetic Calculator. Once you have done this, use the results of the Punnett squares to help you look for the genotypes that you will count as recombinants and which you will count as nonrecombinants. Hint: To help you as you search the Large Family pedigrees you may want to export the results of your Punnett squares from the Genetic Calculator and print a copy of these results to have in front of you as you look at the large family pedigrees to count recombinants and nonrecombinants.
- Counting Recombinants and Nonrecombinants
Once you have located a Large Family pedigree with the double heterozygous and double homozygous parents that you determined with the Genetic Calculator function, begin to tally recombinant and non-recombinant offspring by clicking on the arrow buttons at the bottom of the screen.
After counting offspring for one pedigree, choose another useful pedigree to look for recombination. Note that as you tally recombinants and nonrecombinants, a running tally for the total number of offspring that you have counted is shown at the bottom of the screen along with recombination frequency and LOD score. You will need to count offspring from several pedigrees to accumulate data that will help you determine if the trait and probe are linked. Remember that you are looking for a LOD score of 3 or greater, and a recombination frequency of less than 50% as good evidence that the trait and probe are linked. If you are seeing evidence for linkage then you are ready to map the trait to a chromosome.
Conversely, a recombination frequency of 50% indicates that the probe and trait are probably unlinked. This can usually be determined after analyzing just a few pedigrees. If this is the case, choose another probe and try again with another probe! You should find that recombination data using the asr probe indicates that the asr marker is not linked to the ALS gene. Click on the Chromosome View and note that the asr probe is located on chromosome 1, hence you can probably rule out this chromosome as the location of the ALS gene, and you may want to rule out using the other probes on this chromosome in your next search. You may find that you will have to search for recombination with many probes before you find one that is linked to your trait.
To look for linkage with another probe, return to the Large Family function. Click on the New Search button. This time, search the database for pedigrees using the "juva" probe. Be sure to answer each of the three questions to choose database criteria that will favor the pedigrees that you will need to study recombination. Once you have done this, count recombinant and nonrecombinant offspring again. This time you should see evidence for linkage of the juva probe and ALS gene. Follow the next procedure to map the ALS gene to the correct chromosome.
- The Genetic Calculator function can be very useful for helping you determine which pedigrees will be best suited for studying recombination between the trait and marker. This function allows you to specify genotypes in the parents of a full pedigree that show the trait and marker linked to a pair of homologous chromosomes or unlinked on different chromosomes.
- Mapping the Mutation to a Chromosome
Now that you have evidence for linkage, click on the Chromosome View tab at the top of the screen. Genetic maps appear which show loci for markers on five different chromosomes. Notice that the marker "juva" is located on chromosome 21. To position the ALS gene, click and drag the ALS arrow on the upper right side of this window. Drag the ALS arrow to chromosome 21 and position the arrow relative to the juva marker according to the recombination frequency value that you generated with the Large Family pedigrees. For example, if you generated a recombination frequency of 15%, you should move the ALS arrow to a position that is 15 map units (centimorgans) away from the juva locus. You will not know, however, which side of the juva locus to position the ALS arrow until you study linkage between the ALS locus and the other markers on chromosome 21.Notice the other markers on chromosome 21, "ddy" and "utc3." Return to the Large Family function and repeat a database search using probes for each of these markers to better pinpoint the location of the ALS gene. You should find that the ALS gene maps to a position just above the juva marker. See your instructor for the exact location of the ALS gene.
- Printing Your Chromosome Map and Recombination Data
Print a copy of the Chromosome Map that you developed by clicking on the Export Graphic button in the lower right corner of the screen. A new GIF image window will open which you can now use to print your map.To print your recombination data, return to the Large Family view. You can export a single pedigree that you have labeled by clicking on the Export Pedigree and printing from the GIF image window that appears. You can also export all of the data (total number of recombinants, nonrecombinants, recombination frequency, and LOD scores) for each probe that you used by clicking on the New Search button, then clicking on the Export Whole List button. With this function you will open a window with your data that functions as a lab notebook. You can add any comments that you'd like to your data and print a copy of this data as evidence of your gene hunting progress.
- Begin by clicking on the Large Family tab at the top of the screen. Before you view the family pedigrees, you are asked to answer three questions on the expression of the mutant phenotype in a pedigree. You have the option of selected whether the mutant trait is expressed in grandparents, parents, and offspring. The purpose of these three questions is to better define and narrow your search of the Large Family databases to select for those pedigrees that will be most helpful to study recombination.
On Your Own: Mapping other Mutations
Now that you have experienced how PedigreeLab functions to map the location of the ALS gene, you are on your own to be a gene hunter for some of the other mutations in PedigreeLab!
-
Follow the guidelines in the ALS assignment to help you identify the location and approximate locus for each of the following mutations:
- Huntington’s disease (HD)
- Duchenne muscular dystrophy (DMD)
- Werner’s syndrome (WRN)
- Cystic fibrosis (CF)
- Two additional mutations of your choice.
- For each mutation, perform the following:
- Study several Full Pedigrees, then develop a hypothesis to explain the mode of inheritance for the mutation.
- Use the Large Family
pedigree function to identify pedigrees with parents that will produce offspring
that you can use to look for recombination. Think carefully about the mode of
inheritance for the trait, then decide whether to answer yes or no to the three
questions that ask you to define whether the trait is expressed in grandparents,
parents, and offspring before searching the Large Family database.
Use the Genetic Calculator to help you select the correct Large Family pedigrees and decide what genotypes to look for when counting recombinant and nonrecombinant offspring. Once you have selected the correct pedigree, count the number of recombinant and nonrecombinant offspring. Do this until the recombination and LOD data indicate whether the trait and probe are linked or unlinked. If the trait and probe are linked, use the Chromosome View function to assign a position for the mutant gene on the correct chromosome. If the trait and probe are unlinked, then pick another probe to test for recombination and repeat the process until you find a probe that is linked to the trait.
- Print your recombination data for any probes that you used to search for linkage.
- Once you have found the chromosomal location for the mutation, be sure to test all of the other probes on that chromosome that are linked to the mutation so you can better refine and pinpoint the location of the trait relative to the markers on the chromosome. Your goal is to develop as accurate a map as you can. Print your chromosome map when you are done.
- Show your recombination data and chromosome maps to your instructor to see how accurate your mapping experiments were.
Group Exercises
The Human Genome Project is a worldwide initiative to develop genetic maps for the 22 autosomes and two sex chromosomes in humans. One long-term goal of the Genome Project is to identify and determine the nucleotide sequence for an estimated 100,000 genes that are thought to be present in the approximately 3 billion nucleotides of DNA that comprise the human genome. The Genome Project began in 1990 and was targeting as a 15-year project with an estimated cost of $3 billion. Although this project originated in the United States, labs around the world are contributing to the Genome Project. Many new technologies for identifying and sequencing genes have been developed as a result of the project. Because of this, the progress of the Genome Project is actually slightly ahead of schedule and just under budget. One technique being used by Genome scientists involves using RFLP analysis and other similar techniques to develop extensive genetic maps of chromosomes in humans and other animal and plant species such as the nematode Caenorhabditis elegans, Drosophila melanogaster, the common house mouse, Mus Musculus, and the flowering plant Arabidopsis thaliana. Some labs involved in the Genome Project are working on mapping assigned segments of certain chromosomes. Other groups are involved in checking the accuracy of maps completed to date.
The following exercises are designed to have you investigate the accuracy of genetic mapping as simulated in PedigreeLab, and enable you to map an entire chromosome. Work together in a group of four students to complete the following assignments.
- Select two mutant genes that you have not mapped already. Divide your group
into pairs, and have each pair of students work together to identify the chromosome and approximate locus for each of the two mutations. Once each pair has mapped the two genes, compare your results with each other. Did each pair in your group identify the correct chromosome(s)? Why or why not? If you cannot agree on the correct chromosome be sure to go back and review your work until everyone in your group can agree on a correct chromosome for each mutation.
Once you have agreed on a chromosome location, compare the accuracy of the locus that you identified for each mutation. How accurate were the maps when you compared them? How many map units did the two maps differ by? Explain possible reasons for any differences in loci assigned on each chromosome mapped.
- You have been selected as the leading scientist directing a group of scientists responsible for mapping the X chromosome. Pick three other students who can work as your partners to narrow down your hunt to map the X chromosome.
How would you begin? Should you assign each student a mutant gene that you have not yet mapped and have each person study the inheritance for that gene? Or should you work as a group and look for inheritance patterns to first narrow your search for genes that appear to be X-linked? Decide on an approach to use then work together to develop a map of the X that will show all of the X-linked mutations present in PedigreeLab. Because PedigreeLab will show different regions of the X-chromosome depending on the probes you are using, PedigreeLab will not show one complete full length-map of the X. Your group will have to piece together the different maps of the X shown in PedigreeLab to draw one complete map.
Once you have done this, compare your map with another group of students in your class that has worked on the X.
How accurate were the two sets of maps? Were there any differences in genes mapped to the X when the maps were compared? If there were differences, work together with the other group to resolve these differences until you can agree on an accurate and complete map of the X chromosome. Print your map and show it to your instructor for comparison to the correct map.
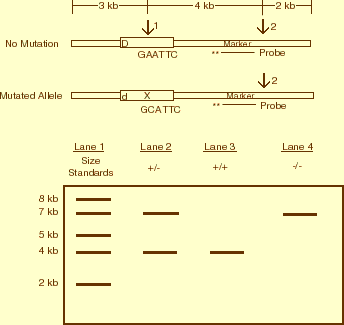
Figure 1
Figure 1: Diagrammatic Representation of
Restriction Fragment Length Polymorphisms (RFLPs) for a Recessive Trait.
Shown is an example of two homologues from an individual who is heterozygous (Dd)
for a recessive gene that is linked to a genetic marker. Arrows indicate the
restriction enzyme recognition sequence that would be cut by restriction
digestion of each chromosome with the enzyme EcoRI. Note two sites (1, 2) are
present on the homologue with the dominant allele (D), while only one site (2) is
present on the homologue with the recessive allele (d). The absence of
restriction enzyme cutting site number 1 in the recessive allele due to a single
nucleotide mutation is indicated with an "X." The location of a radioactive
probe that is complementary to the marker sequence is also shown. The pattern of
restriction fragment length polymorphisms that would be generated for these
homologues is visualized after gel electrophoresis of the digested DNA fragments,
transfer of the DNA fragments to a filter, and hybridization of the fragments to
the radioactive probe, followed by exposing the filter to X-ray film to produce
an autoradiogram. In the autoradiogram, lane 1 shows the migration of DNA size
standards of known length that are used to determine the nucleotide length of
each RFLP. Lane 2, RFLPs for a heterozygote (+/-) where the genotype is
designated according to the presence (+) or absence (-) of a restriction site;
cutting with EcoRI yields two fragments of 7 kb and 4 kb in size. For
comparison, lanes 3 and 4 show RFLPs for individuals who are homozygous for the
restriction site, +/+ or -/-.